Unlock Biobank Collaboration Without Sharing Data
Generate Naive Summary Statistics (NSS) once, enable unlimited GWAS model exploration. Privacy-preserving meta-analysis for the biobank era.
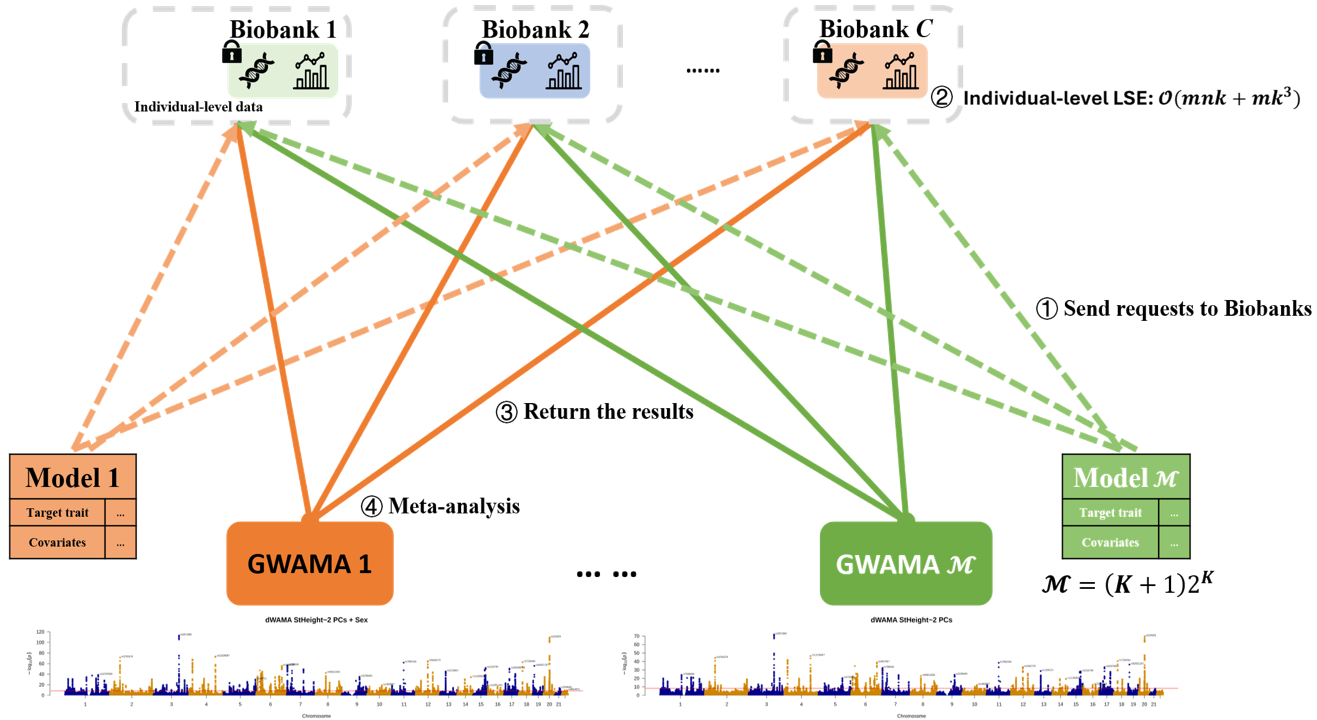
Traditional Meta-Analysis is Broken
Current GWAMA workflows require repeated data requests, limiting model exploration and burdening biobanks with computational overhead.
Traditional GWAMA
- ✗ Request summary stats for each model change
- ✗ Biobanks bear computational burden
- ✗ Only explore 1 of (k+1)2k possible models
- ✗ Weeks of coordination overhead
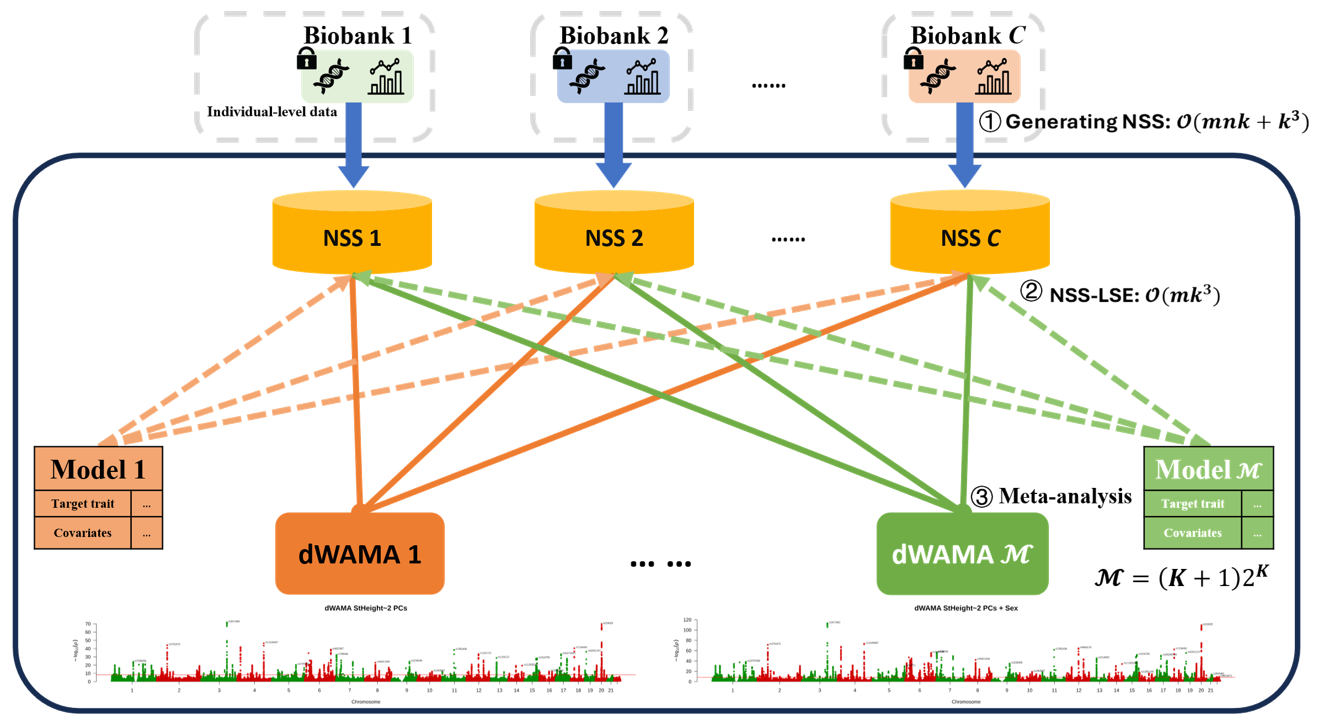
nssgen + dWAMA
- ✓ Generate NSS once, explore all models
- ✓ Computation shifts to researchers locally
- ✓ Access all (k+1)2k covariate combinations
- ✓ Minutes instead of weeks
Built for Modern Genomics
Everything you need to accelerate collaborative genetic research while maintaining the highest privacy standards.
Privacy by Design
Individual-level genetic data never leaves the biobank. NSS contains only variance-covariance matrices—mathematically impossible to reconstruct individual genotypes.
Model Flexibility
Explore all (k+1)2^k possible GWAS models locally. Toggle covariates on/off, swap phenotypes, and fine-tune without re-contacting biobanks.
Computational Efficiency
O(Cmk²) time complexity vs O(Cnmk² + Cmk³) for traditional GWAMA. Analyze 64 models in 2 hours instead of weeks of coordination.
PRS Fine-Tuning
Systematically evaluate covariate effects on polygenic risk scores. Our UK Biobank validation showed R² improvements from 0.02 to 0.05 through model tuning.
Multi-Biobank Ready
Seamlessly combine NSS from multiple biobanks. Validated across 19 UK Biobank cohorts and 4 NIPT biobanks with 149K+ individuals.
API-First Platform
RESTful API for programmatic NSS generation and analysis. Integrate with your existing pipelines, PLINK workflows, and PRS tools.
How nssgen Works
A simple three-step process that transforms how biobanks collaborate on genetic research.
Generate NSS
Biobanks run our tool on their individual-level data once. This produces a compact variance-covariance matrix (NSS) that captures all statistical relationships.
Share Publicly
NSS files are safe to share—they contain no individual-level data. Upload to our platform or distribute through your preferred channels.
Analyze Locally
Researchers download NSS and run unlimited GWAS models locally. No biobank coordination needed. Fine-tune covariates, run meta-analyses, optimize PRS.
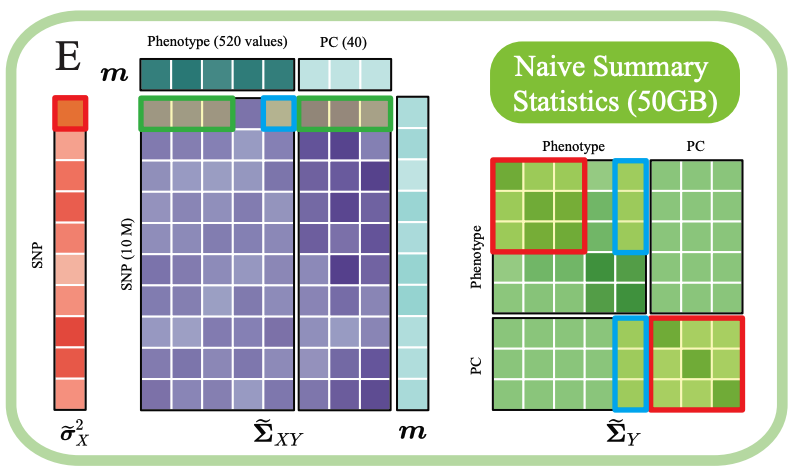
NSS Data Structure
Compact variance-covariance matrices containing phenotypes and principal components. ~50GB for a full biobank.
UKC Computation Engine
Efficient least-squares estimation directly from summary statistics without individual data.
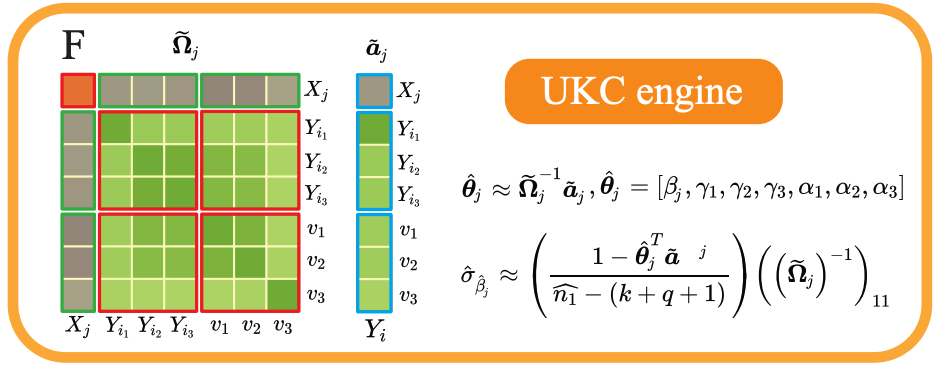
The Math Behind NSS
NSS captures the complete variance-covariance structure needed for any linear regression model. One matrix, unlimited models.
NSS-LSE: Naive Summary Statistics Least Squares Estimation
Proven at Scale
Validated across UK Biobank and real-world NIPT cohorts with biobank-scale sample sizes.
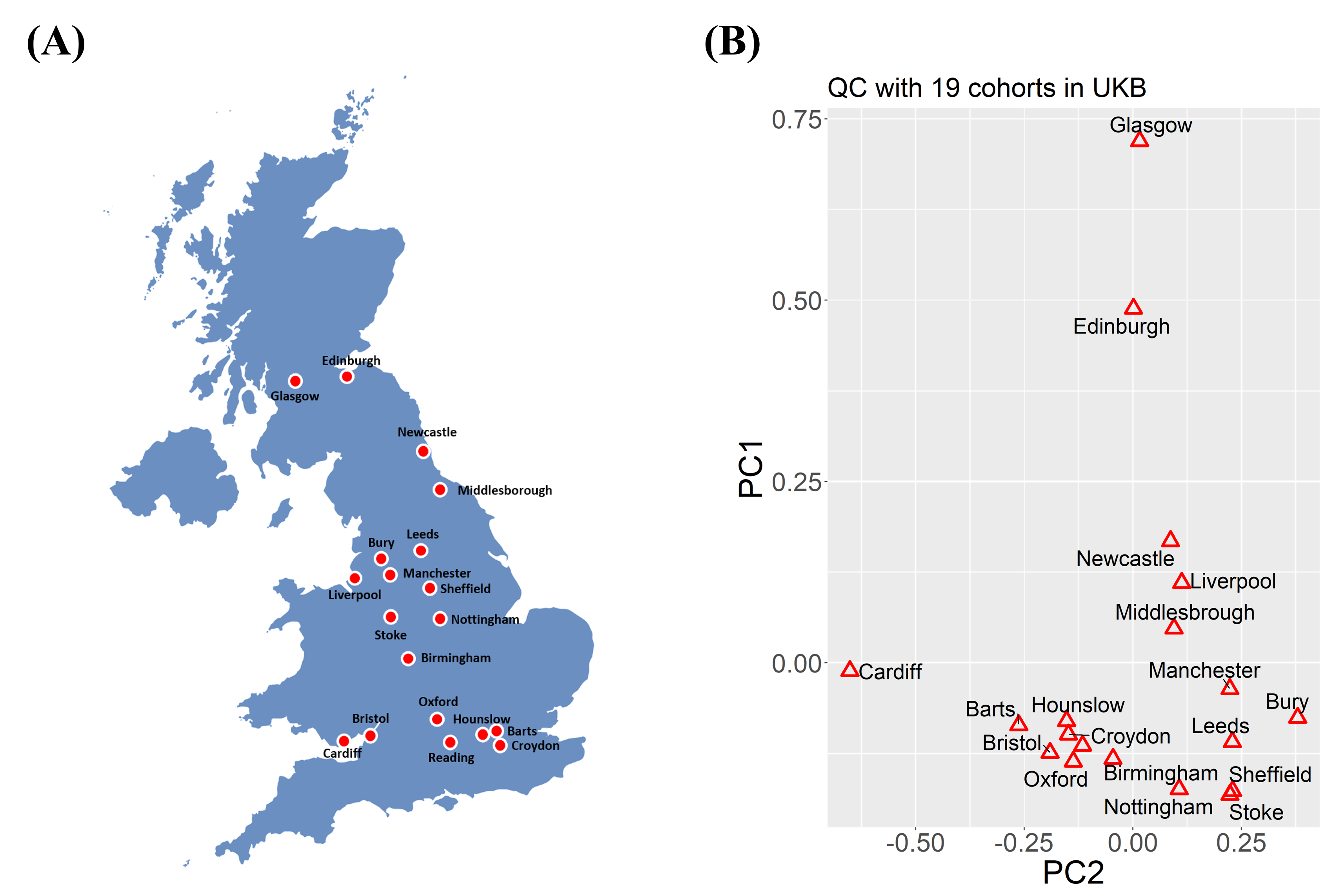
UK Biobank Validation
We validated nssgen across 19 UK Biobank assessment centers, analyzing 280K unrelated white British individuals with 10.5M SNPs.
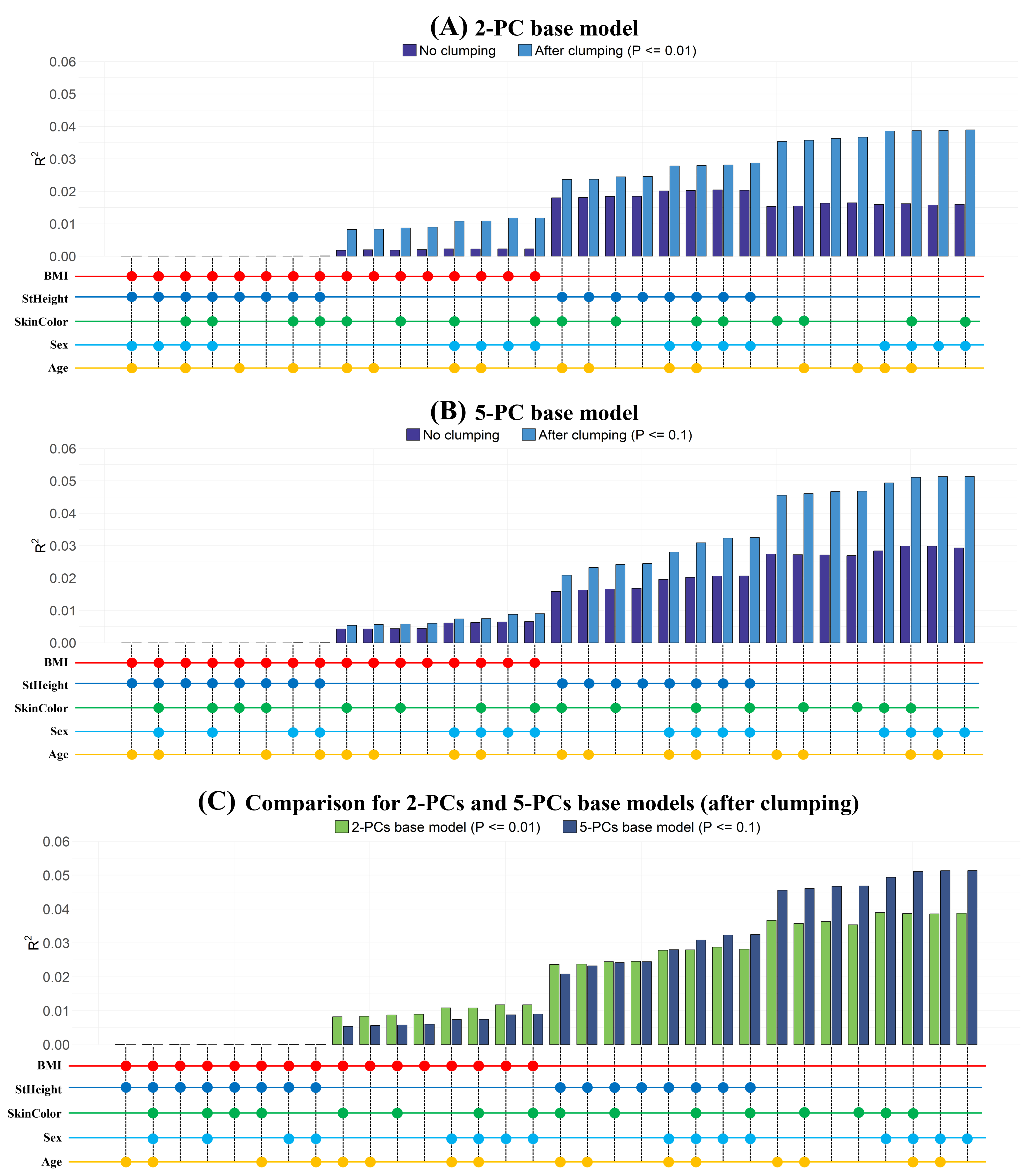
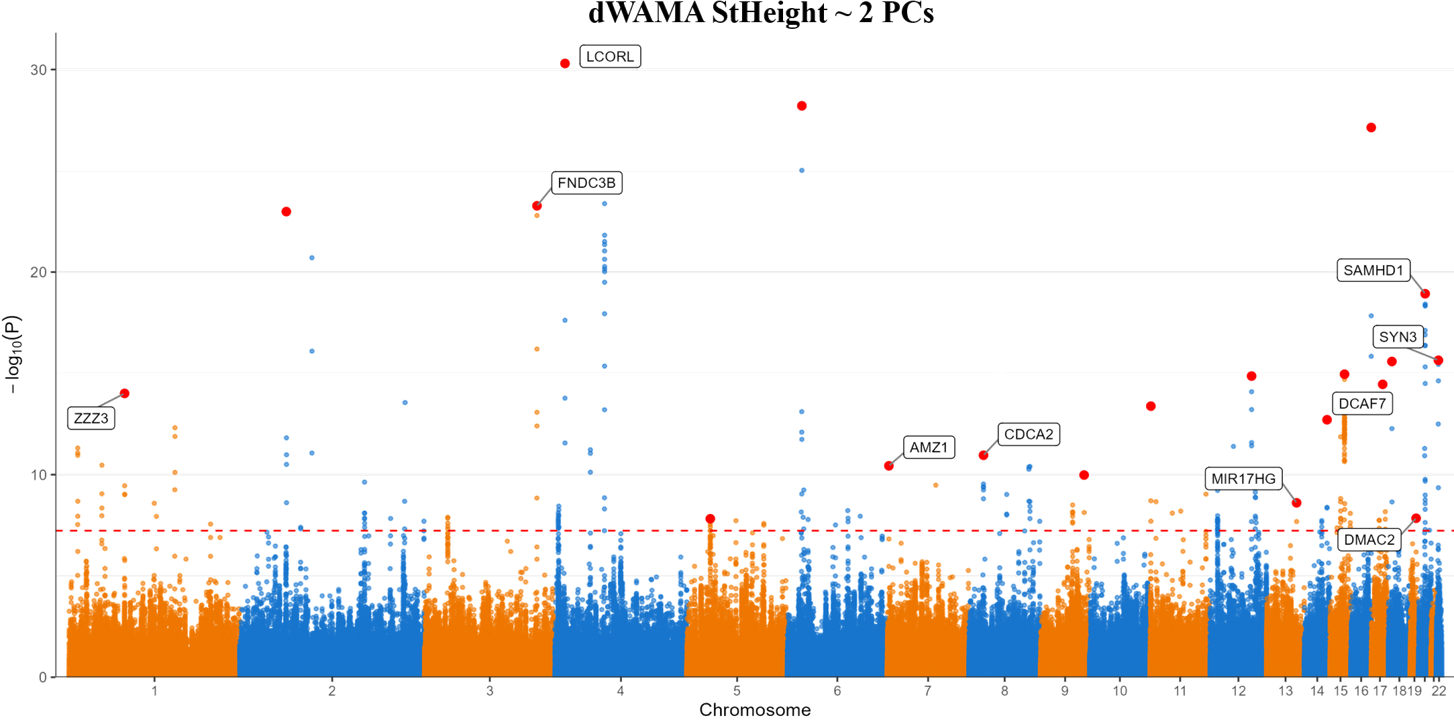
Polygenic Risk Score Fine-Tuning
Systematically explored 64 GWAS models for Weight prediction. nssgen enabled efficient covariate evaluation without repeated biobank access.
Real-World NIPT Cohorts
Validated in a true multi-biobank setting with 4 independent NIPT cohorts from China, demonstrating practical applicability.
nssgen API & Cloud Platform
Generate NSS in the cloud, access pre-computed datasets, and integrate with your existing genomics pipelines through our RESTful API.
- RESTful API for programmatic access
- Pre-computed public biobank NSS datasets
- Python & R SDK packages
- PLINK-compatible output formats
Get Early Access
Join the waitlist for priority access to the nssgen platform and API.
No spam. We'll only email you about nssgen updates.